Join us for conversations that inspire, recognize, and encourage innovation and best practices in the education profession.
Available on Apple Podcasts, Spotify, Google Podcasts, and more.
This online textbook chapter supports and extends the content of the Emerging Infectious Diseases video. The chapter content covers the factors affecting the emergence of disease. Click on the Go button to begin reading, or skip to a sub-section of the chapter in the list below.
During the mid-1900s, most scientists and policymakers were shifting their attention away from infectious disease as vaccines made polio and several other diseases rare, at least in the developed world. Through an intense vaccination campaign, researchers at the World Health Organization (WHO) had eradicated smallpox from the world by the mid-1970s. Most people expected the eradication of other diseases would follow. In the meantime, scientists had created a large array of antibiotics that could easily treat many of the great scourges of history, from leprosy to tuberculosis. Infectious diseases appeared to be on the way out.
This optimistic picture has since changed. Legionnaire’s disease, hantavirus, AIDS (acquired immunodeficiency syndrome), West Nile virus, and SARS (severe acute respiratory syndrome) have rocked the public health and scientific communities. New, drug-resistant strains of bacteria have appeared. Tuberculosis and other old diseases, once thought contained, are again a public health concern. In some of these cases the disease-causing agent was previously undescribed. For others, a previously treatable pathogen somehow changed. In addition, completely new threats emerged. Where had these new threats come from?

Table 1. Factors that affect the emergence of disease
(Smolinski, et al.)
Many factors contribute to the emergence of disease; outbreaks of existing diseases or the emergence of new ones typically involve several of these factors acting simultaneously. Predicting and controlling emerging infection ultimately requires coming to terms with biocomplexity — the elaborate interrelationships between biological systems (including human social systems) and their physical environments.
The human body is inhabited by billions of bacteria. In fact, we normally carry ten times more prokaryotic than eukaryotic cells. Our mouths alone are host to four hundred identified — and probably hundreds more unidentified – species of bacteria. Most bacteria are benign to their host, some even provide valuable services. For example, bacteria in the gut aid digestion and generate vitamins used by their human hosts.
The bacteria we possess are an ecological community; thus, the principles of community ecology and evolution are vital in understanding how these bacteria (both the benign and the potentially harmful) live within us. Each bacterial species is adapted to the habitat and ecological niche it fills, existing in somewhat of an ecological balance. This balance helps thwart the invasion of pathogens, which must compete with resident bacteria for nutrients and space. Resident bacteria also produce antimicrobial proteins called bacteriocins, which inhibit the growth of related species.
When the normal flora are disrupted, it shifts the mix of microbiota and can lead to disease. For example, the use of some broad-spectrum antibiotics can dramatically decrease the numbers of bacteria in the colon. In this situation, Clostridium difficile, normally present only in low numbers, can overgrow. This bacterium produces toxins that cause potentially fatal damage to the lining of the colon. In the few individuals that normally harbor the microbe, normal levels of other bacteria keep C. difficile numbers low. It is only when the balance is disrupted that such a “superinfection” occurs.
Today we face a growing medical crisis: the emergence of bacteria resistant to multiple antibiotics. Strains of at least three potentially fatal bacterial species are now resistant to all the drugs available for treatment. Enterococcus faecalis is generally a benign intestinal bacterium. In the elderly and individuals with compromised immune systems, however, it can be deadly if it gets in the wrong location. E. faecalis can infect heart valves and other organs, causing a deadly systemic disease. Strains of Pseudomonas aeruginosa (which causes skin infections and deadly septicemia) and Mycobacterium tuberculosis (the causative agent of tuberculosis) also evade available drugs. Death rates for tuberculosis have begun to rise, in part because of the evolution of these new strains.
The wide use, and misuse, of antibiotics has encouraged new strains of pathogens to develop. For example, the widespread use of cephalosporin antibiotics has led to drug-resistant E. faecalis. The use of Vancomycin (a drug of last resort) has contributed to the development of VRE (vancomycin-resistant Enterococcus), which defies treatment. Antibiotic-resistant bacteria are generally not more potent and do not generate a more severe disease state; they are, however, more difficult to treat. Resistant bacteria proliferate when a population of microbes containing both susceptible and resistant bacteria are exposed to an antibiotic within the host: susceptible bacteria succumb and resistant bacteria proliferate.
Public health officials urge people to complete the full course of antibiotic treatment. Why? Bacterial susceptibility to an antibiotic is often dose-dependent; an individual bacterium that is only somewhat resistant may survive at low drug concentrations. There will be selection for such more-resistant bacteria and these will eventually predominate the bacterial population. Thus, the failure of patients to complete a full course of treatment, or the use of less than therapeutic doses of antibiotic, can lead to resistant strains. The full course of treatment should be sufficient to wipe out all the pathogenic bacteria.
An additional cause of the rise of antibiotic resistance is the use of antibiotics in animal feed. Humans may be exposed to such as bacteria by handling intestinal contents of the animals, as when butchering or preparing meats. Moreover, bacteria from livestock can get into our water systems.
Various adaptations provide bacteria with antibiotic resistance. Mutations in a target protein that affect binding of an antibiotic to that protein may confer resistance. If an antibiotic inhibits a metabolic pathway and an alternate one becomes available, resistance can occur. Some antibiotic-resistant bacteria make enzymes that destroy drugs; others alter pores in the cell membrane so an antibiotic can no longer enter. Some resistant strains have developed mechanisms for actively pumping antibiotics out of the bacterial cell. The genes for antibiotic resistance are sometimes found on plasmids. The transfer of these plasmids among bacteria facilitates the spread of antibiotic resistance within and between bacterial populations.
The evolution of novel microbes, including antibiotic-resistant strains, depends on diverse members of microbial populations that can thrive in new conditions. Microbes have incredible abilities to change their genetic make-up and evolve faster than their hosts do. Multiple mechanisms ensure the diversity that allows for expansion.
The production of a single, novel gene product may be the key to bacterial survival; however, several gene products working together sometimes provide the advantage. Mutation generates new genes but, unlike higher eukaryotes, bacteria do not undergo sexual reproduction; the typical bacterium simply grows, replicates its DNA, and divides. Therefore, bacterial reproduction does not provide a mechanism for generating progeny with new combinations of genes. How, then, do bacteria obtain new gene assortments, some of which may provide survival?
Bacteria possess several methods for lateral gene transfer (also called horizontal gene transfer), the transmission of genes between individual cells. These mechanisms not only generate new gene assortments, they also help move genes throughout populations and from species to species. The methods include transformation, transduction, and conjugation.
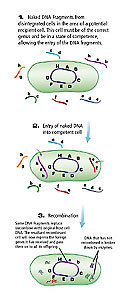
Figure 1. 1. Naked DNA fragments from disintegrated cells in the area of a potential recipient cell. This cell must be of the correct genus and be in a state of competence, allowing the entry of the DNA fragments. 2. Entry of naked DNA into competent cell. 3. Recombination.
Transformation involves the uptake of “naked” DNA (DNA not incorporated into structures such as chromosomes) by competent bacterial cells (Fig. 1). Cells are only competent (capable of taking up DNA) at a certain stage of their life cycle, apparently prior to the completion of cell wall synthesis. Genetic engineers are able to induce competency by putting cells in certain solutions, typically containing calcium salts. At the entry site, endonucleases cut the DNA into fragments of 7,000-10,000 nucleotides, and the double-stranded DNA separates into single strands. The may recombine with the host’s chromosome once inside the cell. This recombination replaces the gene in the host with a variant — albeit homologous – gene. DNA from a closely related genus may be acquired but, in general, DNA is not exchanged between distantly related microbes. Not all bacteria can become competent. While transformation occurs in nature, the extent to which it contributes to genetic diversity is not known.

Figure 2. Flowchart of transduction by bacteriophage.
Transduction is another method for transferring genes from one bacterium to another; this time the transfer is mediated by bacteriophages (bacterial viruses, also called phages) (Fig. 2). A bacteriophage infection starts when the virus injects its DNA into a bacterial cell. The bacteriophage DNA may then direct the synthesis of new viral components assembled in the bacterium. Bacteriophage DNA is replicated and then packaged within the phage particles. Early in the infective cycle the phage encodes an enzyme that degrades the DNA of the host cell. Some of these fragments of bacterial DNA are packaged within the bacteriophage particles, taking the place of phage DNA. The phage can then break open (lyse) the cell. When released from the infected cell, a phage that contains bacterial genes can continue to infect a new bacterial cell, transferring the bacterial genes. Sometimes genes transferred in this manner become integrated into the genome of their new bacterial host by homologous recombination. Such transduced bacteria are not lysed because they do not contain adequate phage DNA for viral synthesis. Transduction occurs in a wide variety of bacteria and is a common mechanism of gene transfer.

Figure 3. The F-pilus serves as a point of contact between a bacterium containing an F-plasmid (the “male”) and a bacterium lacking the plasmid (the “female”). After the female cell is contacted the pilus retracts, pulling the cells together. The exact mechanism of DNA transfer from male to female is not known; it may be by a channel in the pilus or by a temporary fusion of the mating cells.
Some bacteriophages contribute to the virulence of bacterial infections. Certain phages can enter an alternate life cycle called lysogeny. In this cycle, all the virus’s DNA becomes integrated into the genome of the host bacterium. The integrated phage, called a prophage, can confer new properties to the bacterium. For example, strains of Corynebacterium diptheriae, which have undergone lysogenic conversion, synthesize the toxin in diphtheria that damages human cells. Clostridium botulinum and Streptococcus pyogenes, when lysogenized by certain phages, also manufacture toxins responsible for illness, causing botulism and scarlet fever respectively. Strains lacking the prophage do not produce the damaging toxins.
Conjugation is another means of gene transfer in many species of bacteria (Fig. 3). Cell-to-cell contact by a specialized appendage, known as the F-pilus (or sex pilus), allows a copy of an F- (fertility) plasmid to transfer to a cell that does not contain the plasmid. On rare occasions an F-plasmid">single-stranded DNA/ may recombine with the host’s chromosome once inside the cell. This recombination replaces the gene in the host with a variant — albeit homologous – gene. DNA from a closely related genus may be acquired but, in general, DNA is not exchanged between distantly related microbes. Not all bacteria can become competent. While transformation occurs in nature, the extent to which it contributes to genetic diversity is not known.

Figure 2. Flowchart of transduction by bacteriophage.
Transduction is another method for transferring genes from one bacterium to another; this time the transfer is mediated by bacteriophages (bacterial viruses, also called phages) (Fig. 2). A bacteriophage infection starts when the virus injects its DNA into a bacterial cell. The bacteriophage DNA may then direct the synthesis of new viral components assembled in the bacterium. Bacteriophage DNA is replicated and then packaged within the phage particles. Early in the infective cycle the phage encodes an enzyme that degrades the DNA of the host cell. Some of these fragments of bacterial DNA are packaged within the bacteriophage particles, taking the place of phage DNA. The phage can then break open (lyse) the cell. When released from the infected cell, a phage that contains bacterial genes can continue to infect a new bacterial cell, transferring the bacterial genes. Sometimes genes transferred in this manner become integrated into the genome of their new bacterial host by homologous recombination. Such transduced bacteria are not lysed because they do not contain adequate phage DNA for viral synthesis. Transduction occurs in a wide variety of bacteria and is a common mechanism of gene transfer.

Figure 3. The F-pilus serves as a point of contact between a bacterium containing an F-plasmid (the “male”) and a bacterium lacking the plasmid (the “female”). After the female cell is contacted the pilus retracts, pulling the cells together. The exact mechanism of DNA transfer from male to female is not known; it may be by a channel in the pilus or by a temporary fusion of the mating cells.
Some bacteriophages contribute to the virulence of bacterial infections. Certain phages can enter an alternate life cycle called lysogeny. In this cycle, all the virus’s DNA becomes integrated into the genome of the host bacterium. The integrated phage, called a prophage, can confer new properties to the bacterium. For example, strains of Corynebacterium diptheriae, which have undergone lysogenic conversion, synthesize the toxin in diphtheria that damages human cells. Clostridium botulinum and Streptococcus pyogenes, when lysogenized by certain phages, also manufacture toxins responsible for illness, causing botulism and scarlet fever respectively. Strains lacking the prophage do not produce the damaging toxins.
Conjugation is another means of gene transfer in many species of bacteria (Fig. 3). Cell-to-cell contact by a specialized appendage, known as the F-pilus (or sex pilus), allows a copy of an F- (fertility) plasmid to transfer to a cell that does not contain the plasmid. On rare occasions an F-plasmid may become integrated in the chromosome of its bacterial host, generating what is known as an Hfr (high frequency of recombination) cell. Such a cell can also direct the synthesis of a sex pilus. As the chromosome of the Hfr cell replicates it may begin to cross the pilus so that plasmid and chromosomal DNA transfers to the recipient cell. Such DNA may recombine with that of its new host, introducing new gene variants. Plasmids encoding antibiotic-resistance genes are passed throughout populations of bacteria, and between multiple species of bacteria by conjugation.
Lateral gene transfer is a potent evolutionary force that can create diversity within bacterial species (See the Microbial Diversity unit.) As genes for virulence factors and antibiotic resistance spread between and among bacterial populations, scientists are realizing how integral these mechanisms are to the emergence of novel pathogens.
Transposons (transposable elements) are genes that can move (“jump”) from one DNA molecule to another in a cell, or from one location to another on the same DNA molecule. They can facilitate the transfer of genes, such as antibiotic-resistance genes, from the chromosome of a bacterium to a plasmid. They also can contribute to genetic diversity by causing mutations.
The simplest type of transposon is an insertion sequence (IS). It is a sequence of DNA that encodes an enzyme called transposase, which enables the IS to move. The transposase gene is flanked on either side by fifteen to twenty-five base pairs, arranged as “inverted repeats.” A composite transposon is composed of any gene sandwiched between two IS sequences; this entire unit will move.
Bacteria move readily from person to person; global travel has contributed significantly to the dissemination of novel pathogens, including drug-resistant strains. Stuart Levy refers to antibiotics as “societal drugs.” They not only affect the bacteria in a treated individual, but also produce long-lasting changes in the kinds and proportions of bacteria in the environment and in human populations at large. For example, the multidrug resistant Streptococcus pneumonia (a bacterium that causes pneumonia and meningitis) has migrated from Spain to the United Kingdom, the United States, and South Africa.
Crowding also contributes to the dissemination of novel pathogens. Hospitals and nursing homes are particularly ideal environments for the exchange of microbes, including drug-resistant strains. Every year two million people acquire infections while hospitalized and 77,000 people die. Healthy caregivers and visitors can be unwitting carriers, but the scenario is worsened by the compromised status of patients. Cancer treatments and other immunosuppressives, such as those used for transplant patients, contribute to the problem. So does HIV. But any procedure, such as surgery or catheterization, that breaches the protective barrier of the skin increases the risk of infection. In crowded cities, especially in third world countries where adequate sanitation may be lacking, microbes arrive by immigrants from diverse locations. These bacteria can spread rapidly, particularly when immunizations and health care are unavailable.
The evolution of new pathogens is not just a function of human-pathogen or human-human interactions. Sometimes people also unwittingly provide new environments where disease-causing organisms thrive. In the 1970s, for example, air-conditioning systems became widely available. A bacterium normally found in fresh water lakes, Legionella pneumophila, moved into the systems, gaining access to susceptible humans. The result was a previously unreported respiratory infection.
Scientists have identified more than one hundred species of pathogenic bacteria that can infect both humans and animals. As you might imagine, zoonoses(diseases that can be transmitted to humans from other vertebrate hosts) are harder to eradicate. For example, Lyme disease is a zoonosis that has emerged, in part, because of human alteration of ecosystems. (See the Biodiversity unit.) A recent example of a probable zoonosis is SARS, which has been found in the civet cat and other animals.
Influenza
An average of about 36,000 Americans die each year as a result of influenza. The “Spanish Flu” of 1918-9 killed more people worldwide than did World War I. This disease involves the interaction of multiple animal hosts; however, the story is more complicated. Variation among influenza viruses occurs at the level of the hemagglutinin (HA) and neuraminidase (NA) spikes, which cover the viruses’ outer envelopes. These proteins are important for the attachment, and eventual release, of the virus from host cells. In response to an infection the immune system mounts a response against these proteins. Nonetheless, an individual immune to one subtype of influenza may not be able to mount an immune response to a new subtype with modified hemagglutinin or neuraminidase. Genetic mutations, resulting from the change of one or more amino acids within HA or NA, are responsible for the recurrence of minor epidemics of influenza in two- to three-year cycles. This is referred to as antigenic drift.

Figure 4. A new form of a virus can arise when multiple viruses infect the same animal cell. Segments of nucleic acid can be exchanged, resulting in a novel pathogen.
More dramatic changes, called antigenic shifts, occur when multiple viruses cause coinfections in animal cells (Fig. 4). For example, aquatic birds serve as reservoirs for the influenza-A virus. Some, but not all, types of bird influenza can infect humans directly. Occasionally, a new form of the virus – a new human pathogen – arises when multiple viruses infect the same cell. The mixing vessel is often the pig, which can be infected by both the bird and human forms of the virus. Influenza is an RNA virus and its genome is oddly segmented. Genes for HA and NA are found among the eight distinct fragments of single-stranded RNA. If a pig cell is infected with viruses from two different sources, RNA segments might be exchanged. Such genetic exchange can dramatically change the nature of the spikes found on the newly derived virus. Major pandemics of influenza, including the 1918 flu and the “Hong Kong” flu of 1968, have occurred immediately after antigenic shifts have taken place. Farms and markets where poultry, pigs, and humans come in close contact are considered important to the emergence of new subtypes of influenza.
Lyme disease and influenza are just two examples of diseases that have emerged because of human contact with animal reservoirs. Understanding the epidemiology of other emerging infections, such as hantavirus and ebola, also depends on an understanding of animal hosts.
Insects provide a system that can deliver pathogens directly to the bloodstream and are essential to the spread of some infections. From a pathogen’s perspective, moving from host to host is essential to survival; yet, the skin presents a barrier. Wounds, burns, and catheters provide opportunities for entry for some pathogens, but insect-borne bacteria have an advantage. Still, arthropod-transmitted microbes must be able to survive in the arthropod’s gut, proliferate, and then become positioned (such as in the insect’s salivary gland) for delivery to the animal host.
Malaria
Members of the protozoal species Plasmodium, which cause malaria, have evolved a successful relationship with their

Figure 5. Sporozoites are delivered to the human bloodstream from the salivary gland of the Anopheles mosquito when the insect bites. In the liver, the sporozoites multiply and become merozoites. The merozoites enter red blood cells and become trophozoites. Red blood cells rupture and new merozoites, which have developed from the trophozoites, are released. Gametocytes (the sexual stage) are eventually produced. Gametocytes taken up by the mosquito in a blood meal fuse to form zygotes, which give rise to sporozoites.
arthropod vector, the Anopheles mosquito. Malaria is prevalent in areas where this mosquito thrives — in parts of Africa, Asia, and China. Three million people die every year from Plasmodium infections. Between 1950 and 1970, efforts to eradicate malaria involved the use of the insecticide DDT. Unfortunately, mosquitoes developed resistance to the spray. Now considered a reemerging disease, malaria incidence is on the rise as eradication programs failed and drug-resistant strains of the parasite have evolved. The complex life cycle of the parasite makes development of vaccines difficult, and efforts to reduce malaria by controlling its insect vector continue (Fig. 5).
Dengue
Approximately eighty viruses depend on insects for transmission. The virus that causes dengue and dengue hemorrhagic fever has the broadest distribution, comparable to that of malaria. Approximately 2.5 billion people live in areas at risk for dengue, and millions are afflicted each year. The fatality rate is about five percent, with most fatalities occurring in children and young adults. Transmitted by the mosquito Aedes aegypti, dengue, or “breakbone fever,” causes a range of symptoms: nausea and weakness, severe bone and joint pain, and high fever.

Distribution of the mosquito Aedes aegypti, the vector for dengue/dengue hemorrhagic fever. A mosquito eradication program administered by the Pan American Health Organization ended in 1970.
Four immunologically distinct types of the virus exist, so individuals can contract the disease four times during their lifetime. An infection with a second subtype of the virus may result in a severe hemorrhagic disease, involving leakage of blood or fluid from mucous membranes. The hemorrhage seems to involve an immune reaction, resulting from sensitization in a previous infection. A global pandemic of dengue began in southeast Asia after World War II. In the 1980s dengue hemorrhagic fever began a second expansion into Asia, with epidemics in Sri Lanka, India, the Maldive Islands and, in 1994, Pakistan. During the 1980s epidemic dengue arose in China, Taiwan, and Africa. Aedes aegypti and an alternate mosquito vector, Aedes albopictus, are present in the United States. Two outbreaks of dengue were reported in Texas during the 1980s, which were associated with epidemics in northern Mexico.
The dramatic global emergence of dengue relates in part to the lack of effective mosquito control in afflicted countries. Often, deteriorating public health infrastructures are to blame.
Arthropods, important in the spread of many diseases, are particularly sensitive to meteorological conditions. Anopheles mosquitoes, for example, only transmit malaria where temperatures routinely exceed 60° F. Temperature influences the proliferation rate of the mosquito, as well as the maturation rate of the parasite within the insect. Mosquitoes live only a few weeks; warmer temperatures raise the odds that the parasites will mature in time for the insect to spread the protozoans to humans.
Global climate change has already altered the species ranges of a number of animals and plants. (See the Biodiversity unit.) Further change may increase the range of the mosquito vectors that transmit disease. This could expose sixty percent of the world’s population to malaria-carrying mosquitoes. (Forty-five percent of the human population now reside in a zone of potential malaria transmission.) In fact, malaria is reappearing in areas north and south of the tropics, including the Korean peninsula and areas of Europe. During the 1990s outbreaks of locally transmitted malaria occurred in Texas, Florida, Georgia, Michigan, New Jersey, New York, and Ontario. Although these incidents probably started with a traveler or stowaway mosquito, conditions were such that the infection could be transmitted to individuals who had not been traveling.
Cholera and Global Climate Change
Global climate change may also bring flooding. In addition to creating breeding grounds for insects, this could increase the incidence of water-borne diseases such as cholera. The bacterium Vibrio cholerae causes seasonal outbreaks of intestinal infection so severe that individuals can lose as much as twenty-two liters (six gallons) of fluid per day. The intestinal lining becomes shredded so that white flecks, resembling rice grains, are passed in feces. Without adequate fluid replacement, death can occur in hours. During a 1991 epidemic in Bangladesh 200,000 cases were counted in only three months.
Historically, cholera (caused by V. cholerae) has been a problem in coastal cities, especially those where the quality of the water supply is poor. In an 1849 groundbreaking study, John Snow mapped cholera deaths in London and realized that victims had been drinking from the same well. The association between cholera and contaminated water was established, and appropriate water treatment seemed to bring the threat under control. Yet, especially in areas where water treatment is unaffordable, cholera epidemics continue.
Where does Vibrio cholerae go between epidemics? This question intrigued Rita Colwell and her associates. Surprisingly, they found the bacterium in Chesapeake Bay in a dormant, spore-like form that was difficult to culture in the laboratory. Colwell used antibodies, directed to a component of the bacteria’s cell membrane, and was able to detect the dormant organism. In this form V. cholerae survives in a range of habitats, including seawater, brackish water, rivers, and estuaries. Colwell also found that wherever tiny crustaceans known as copepods were abundant so were the bacteria, which cling to the copepod and colonize its gut.
Understanding the reservoir for cholera may be important to unraveling the periodicity of epidemics. Colwell turned her attention to locations where cholera outbreaks were common, such as in Bangladesh. By reviewing data from satellite monitors, she noticed that seasonal peaks in sea-surface temperatures in the Bay of Bengal correlated with the number of cholera admissions in nearby hospitals. Similar correlations existed between sea-surface temperatures and South American cholera epidemics in the 1990s. It is possible that the rise in temperature raises sea-surface height, driving seawater into estuaries. Alternately, rising temperatures might provide the right set of environmental conditions to boost copepod populations, perhaps by increasing populations of the photosynthetic plankton, which copepods feed upon. In either case, recognizing the association between sea-surface temperature and cholera incidence may make epidemics easier to predict. The relationship between climate and epidemic also increases the concerns raised by global climate change.
Climate and Hantavirus
Weather patterns can also influence the numbers of vertebrate animals serving as reservoirs for human pathogens. In 1993, in the Four Corners area of the United States (where New Mexico, Arizona, Utah, and Colorado meet), researchers tracked an outbreak of pulmonary illness that killed half of those infected. The causative agent, hantavirus, was not a new threat but was endemic in the rodent population of the area. Researchers were able to find the deadly virus in mouse tissue archived years earlier. Hantavirus spreads to humans by rodent urine and droppings. During the mild, wet winter of 1993, piñon nuts, a favored food for the deer mouse, flourished. As rodent populations soared, the opportunities for mouse-human interactions increased. Native American legend describes an association between piñon nut abundance and illness. Scientists found an association between the periodic climate pattern El Niño-Southern Oscillation and outbreaks of hantavirus.
Medical practices, the adaptability of microbes, global travel, crowding, human susceptibility, alternate vertebrate hosts, insect vectors, and climate are just some of the factors that influence the emergence of disease. In most cases the interplay between multiple factors must be understood. Not the least of these is deteriorating public health systems in many countries where substandard water and waste management continues. War and famine also set up conditions that lead to the emergence of disease and, especially in poor nations, the political impetus to implement prevention and control strategies is often lacking.
The prevention and control of emerging infectious diseases requires a global perspective that accounts for biocomplexity, all the interrelated factors that contribute to the evolution and survival of infectious agents. Individuals from many disciplines – biologists, chemists, statisticians, atmospheric scientists, and ecologists – must work together. Effective surveillance is essential. Multiple control measures will often be appropriate. New genomic and proteomic techniques may provide not only more effective detection but also prevention by novel vaccines.
The effective interaction between public health officials and individuals from a variety of disciplines was exemplified during the West Nile virus outbreak that occurred in the New York City area during the summer of 1999. By mid-October forty-eight people had demonstrated an unusual illness characterized by fever, extreme muscle weakness, and pneumonia-like symptoms. Four had died. Encephalitis or meningitis was present in a few of the more serious cases. West Nile virus was identified as the causative agents using antibody-based tests and DNA comparisons.
West Nile virus hails from Africa, Australia, and the Middle East, and had never been seen in the Western Hemisphere. At the time city wildlife officials and veterinarians at the nearby Bronx zoo were struggling with a peculiar infection among crows and the zoo’s collection of exotic birds. Brain hemorrhages and heart lesions were observed in dissected birds; DNA analysis showed the presence of West Nile virus. The discovery of the virus in wild bird populations, which could potentially serve as a reservoir for human disease, spawned surveillance of birds throughout the United States. Concerns that migratory birds would spread the virus rose. Flocks of chickens were used to monitor viral spread. In the meantime New York City began spraying for mosquitoes. By September 2002 the virus had infected a woman in Los Angeles. Continued surveillance of the bird population and continuing communication between wildlife experts, public health workers, and others will be instrumental in curtailing this infection in the United States.
Effective surveillance is a critical step in preventing the spread of emerging diseases. For example, the new influenza vaccine available each year is the result of constant vigilance. The World Health Organization and others identify the strains of influenza most likely to cause infection in the coming year and define the vaccine based on their findings. In this case an understanding of the animal reservoirs of the disease is important to the surveillance effort. The emergence of novel strains is most likely where poultry, pigs, and humans come in close contact. As a result, monitoring is conducted where such conditions abound.
It is often necessary to take multiple measures to control disease. In the case of malaria the first steps to prevention are as simple as the use of bed nets for reducing bites from mosquitoes and more frequent draining of flooded environments (such as rice fields) where mosquitoes thrive. In the end, DNA-based vaccines, founded on an understanding of the complex life cycle of the protozoal parasite, may be the answer.
Daniel Carucci of the U.S. Naval Medical Research Center and others have identified various proteins that are expressed by the malarial parasite during different stages of its life cycle. Some of these proteins should be recognized as foreign by the immune system and might serve as vaccines. The goal is to stimulate the production of not only antibodies but also cellular immunity specific for various stages of the parasite. (See the HIV and AIDS unit for an introduction to the immune system.) Rather than injecting the proteins into individuals, Carucci is evaluating the use of DNA vaccines. Such vaccines usually comprise DNA, encoding the protein(s) of interest, adsorbed onto gold particles and injected with an air gun into muscle tissue. The expression of malarial proteins by recipient cells and the subsequent immune response to the proteins is being evaluated. If successful, DNA-based vaccines might offer advantages over traditional vaccines; they are less expensive to prepare and easier to store than protein-based vaccines. However, DNA can serve as an immunogen itself; it is thought that diseases such as lupus result from an immune reaction to DNA. As vaccine development continues, the importance of traditional public health measures to prevent and treat malaria remains essential.
The threat from established and evolving disease organisms remains with us. Given high reproductive rates and mechanisms for lateral gene transfer, microbes can adapt to and rapidly circumvent the best treatments scientists develop. We have seen how new diseases arise and spread when humans interact with each other or with the environment in new ways. The anthrax attacks in the fall of 2001 remind us that the threat of bioterrorism continues. This ancient form of warfare dates as far back as 1346 when the Tartar army catapulted the bodies of plague victims into the city of Kaffa.
The journalist Laurie Garrett has suggested that because human behavior influences the emergence of disease, we have significant control over our struggle with microbes. Certainly, our understanding of the factors that contribute to the evolution of new pathogens is continuing to increase, and experience with such outbreaks as West Nile has helped hone surveillance and control measures. However, the global nature of disease means that public health strategies must be global as well.